Narrative Information Access for Precise and Structured Literature Searches
Summary:
The essential component of scientific discourse is the publication of knowledge in stringent lines of arguments. Digital libraries curate these publications in extensive collections and must provide effective access paths to them. However, searching for arguments with keywords is often not precise enough. In close cooperation between the University Library and the Institute for Information Systems at the Technische Universität Braunschweig, a narrative service is being developed in the Specialized Information Service Pharmacy (in German Fachinformationsdienst Pharmazie) to provide a precise and variable-based search for targeted lines of arguments in scientific literature. The service will be integrated into the central service – the drug-centered search platform PubPharm (www.pubpharm.de). The narrative service offers a precise search for lines of arguments by allowing users to formulate their queries as relationships between pharmaceutical entities (such as active ingredients, diseases, targets). For example, pharmaceutical mechanisms can be formulated and precisely searched. In addition, the service supports placeholders in the queries that allow users to explore their research field. For example, the service can visualize all drug-disease relationships with associated literature in a structured representation. The narrative service requires a pre-processing of documents, i.e., annotating pharmaceutical entities and extracting relationships between them. The pharmaceutical community is currently evaluating the narrative service. In this paper, we present the technique of our narrative service and discuss its transferability to other domains of various disciplines.
Zusammenfassung:
Der essentielle Bestandteil des wissenschaftlichen Diskurses ist die Veröffentlichung von Wissen in stringenten Argumentationen. Digitale Bibliotheken kurieren diese Veröffentlichungen in großen Sammlungen und bieten Zugangspfade für das Wissen an. Die Suche nach Argumenten mit Schlüsselworten ist jedoch oft nicht präzise genug. In enger Kooperation zwischen der Universitätsbibliothek und dem Institut für Informationssysteme der Technischen Universität Braunschweig entsteht im Fachinformationsdienst (FID) Pharmazie ein narrativer Service, um eine präzise und variablen-behaftete Suche nach gezielten Argumentationsstrukturen in wissenschaftlicher Fachliteratur zur Verfügung zu stellen. Dieser wird im zentralen Dienst des FID Pharmazie – der wirkstoffzentrierten Rechercheplattform PubPharm (www.pubpharm.de) – zur Verfügung gestellt. Der narrative Service bietet eine präzise Suche nach Argumentationsstrukturen, indem Nutzer*innen gezielt Anfragen nach Interaktionen zwischen relevanten pharmazeutischen Entitäten (wie z.B. Wirkstoffe, Krankheiten, Targets) formulieren können. Beispielsweise können so zentrale Wirkmechanismen formuliert und präzise gesucht werden. Ergänzend wird eine variablen-behaftete Suche bereitgestellt. Hier können Nutzer*innen ihr Forschungsfeld explorieren, indem beispielsweise sämtliche Wirkstoff-Krankheit Beziehungen mit zugehörigen Referenzen strukturiert visualisiert werden. Um solche Suchen zu ermöglichen, werden Dokumente vorverarbeitet, indem wichtige Entitäten annotiert und Interaktionen zwischen diesen extrahiert werden. Der narrative Service wird aktuell von der pharmazeutischen Fachcommunity evaluiert. In diesem Beitrag wird die Technik des narrativen Service vorgestellt und die Übertragbarkeit auch für andere Domänen unterschiedlichster Fachdisziplinen erörtert.
1.Introduction
In close cooperation between the University Library and the Institute for Information Systems at the Technische Universität Braunschweig, the drug-centered search platform PubPharm (www.pubpharm.de) is being developed to assist the pharmaceutical research community.1 PubPharm provides a comprehensive set of more than 55 million pharmacology-, chemistry- and pharmacy-specific publications: journal articles, preprints, information on clinical trials, subject-specific patents, books, e-books, and dissertations. The location-based availability check supports users to access many electronic resources directly. In addition to a classical keyword-based search, PubPharm offers a structure search capability, including similarity and substructure search. Search results may be linked to relevant pharmaceutical and bioinformatical sources, e.g., KEGG2, Uniprot3, ChEMBL4, and DrugBank5. PubPharm must provide effective and efficient access paths for users to navigate through these extensive collections.6 On the one hand, a service must retrieve precise results to answer a user’s query quickly. On the other hand, a service should generate structured literature overviews to display current trends and the latest research. Traditional keyword-based search engines can only guess the user’s intent, e.g., the relationships between different keywords. The central idea for our service is that users can formulate their information need as a short narrative.
A narrative is basically a short story of interest, i.e., relationships between central entities. On the one hand, such a narrative supports a precise literature search by stating the relevant relationships explicitly. For example, a narrative query might ask for documents that describe a drug-disease treatment like metformin treatments in diabetic patients. Then, the system can precisely retrieve documents that support the queried statement. On the other hand, variables can be used in queries to ask for structured literature overviews, e.g., searching for documents that describe some drug to treat diabetic patients. Then, our system retrieves corresponding documents that meet the user’s information need. Such narrative queries may support researchers with entity-centric literature overviews to structure and navigate through today’s extensive collections. Here, the system aggregates the documents by the corresponding drug substitution, e.g., one group of documents may include the drug Metformin, and another group may include Repaglinide.
What are the requirements for a narrative service? First, natural language texts must be transformed into a structured representation. Usually, knowledge extraction from text requires domain-specific training examples to train suitable extraction models. Therefore, we built upon open information extraction and designed algorithms that close the missing gaps for practical applications. We published our toolbox for the nearly unsupervised extraction of knowledge from texts.7 The toolbox’s code is available as open source, and the software can freely be used or adapted8. Researchers from other domains may use our toolbox to transfer the narrative service to their field of interest. In addition to information extraction, our service requires two domain-specific vocabularies: an entity vocabulary comprising the entities of interest and a relation vocabulary containing the relevant relations between these entities. We introduced the narrative service and reported on evaluation results before9. In contrast, this article focuses on technical aspects and the transferability of the narrative service. The Specialized Information Service Pharmacy will share new insights, services, and research with the digital library community.
This article is structured as follows: We describe our narrative model in Sect. 2, discuss the narrative extraction and retrieval in Sect. 3, and describe relevant implementation details about our prototype in Sect. 4. Finally, we discuss how to transfer the service to another domain in Sect. 5 and conclude in Sect. 6.
2.Narrative Model
Nowadays, the scientific discourse is based on exchanging knowledge. Knowledge might be written down in publications, collected as research data, or gathered in extensive knowledge repositories like Wikidata.10 However, making sense out of these vast amounts of knowledge is still challenging. On the one hand, the processing of unstructured natural language texts is cost-intensive and requires domain-specific expertise. On the other hand, inferring new knowledge by utilizing structured repositories requires identifying relevant and meaningful patterns.
Therefore, we designed a conceptual model, called ‘narrative overlay’, which allows users to formulate their scientific discourse (see Figure 1 for an overview).11 The model is designed as a logical overlay on top of different knowledge repositories. Thus, knowledge of various sources can be combined to validate a possible discourse in the sense of evidence. However, the model is conceptual, and many questions are still open. To understand whether a scientific discourse has some evidence, we introduced so-called ‘narrative bindings’. A narrative binding takes a narrative’s relationship and binds it to a knowledge repository. Such a binding gives evidence about the relationship, e.g., the statement that Metformin treats diabetes mellitus might be bound against a clinical trial or a specialized database. However, if narrative bindings are computed against several knowledge repositories, the context of information must also be considered in that process. As a prime example, biomedical knowledge is usually based on some conditions, e.g., a successful treatment is only observed for patients taking a drug in a sufficient dose and time interval. Therefore, such context information must be considered when combining knowledge of different sources. In another study, we found that fusing information without considering the context had a very negative impact on the results.12 We plan to do more research in that direction.
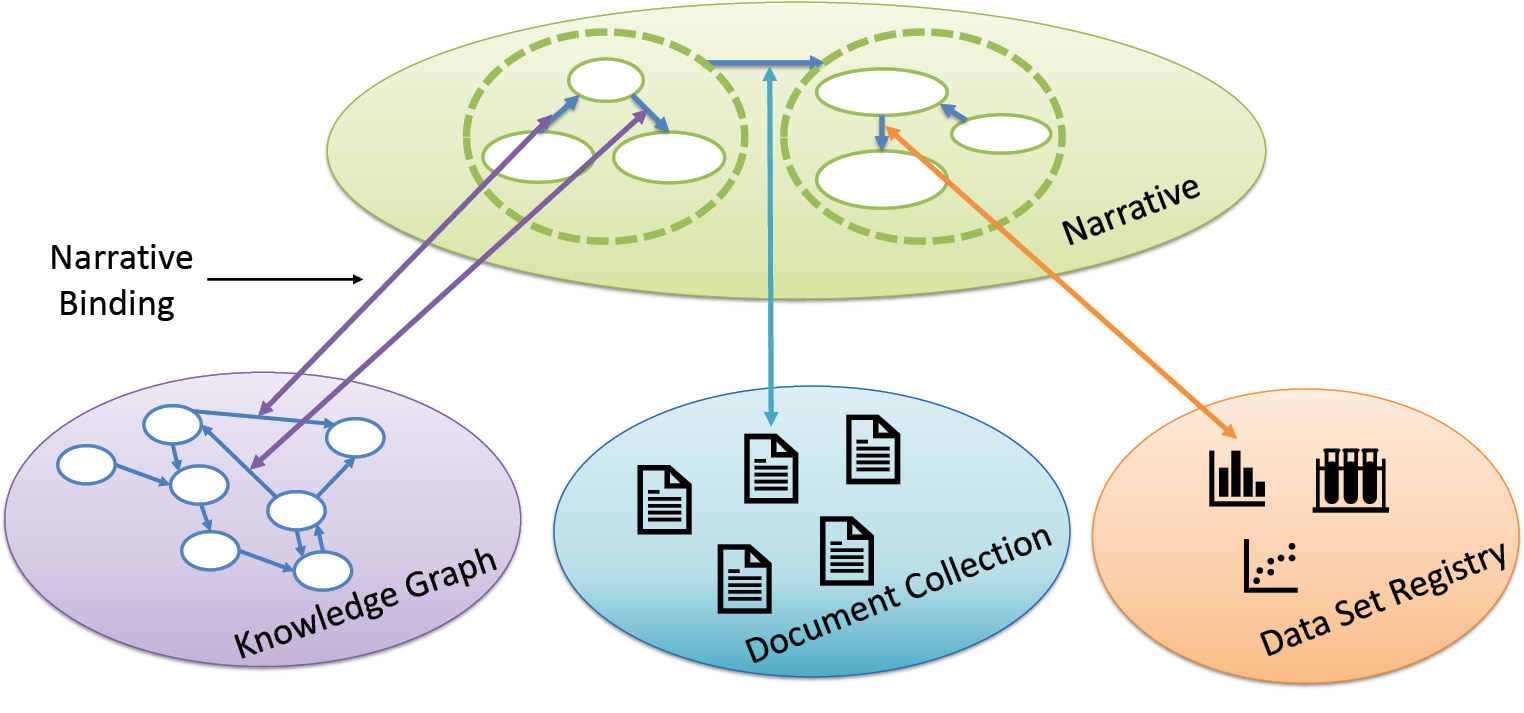
For now, our narrative service builds upon this narrative model by using a suitable subset. We call that subset ‘narrative query graphs’. Users may formulate their information need as a short narrative consisting of entities and relationships between them. Hence, a narrative query graph is a set of fact patterns. Each fact pattern asks for a single relationship between two entities. In addition, we allow users to replace any entity with a variable that may be typed. For example, users might query for drugs that are used to treat diabetes mellitus in humans. The query can be written as: ?X(Drug) treats diabetes mellitus. Then, our narrative service answers this query by matching it against our document collection. If a document matches the query, i.e., all statements of the query are included in the document, it is displayed to the user. Currently, the service matches the query against each document graph one by one. Hence, the query is answered within the context of a single document. However, a complex scientific discourse might only be supported by combining several documents, e.g., one document supports the theoretical background while another document gives empirical evidence. Combining these documents to answer a single narrative query graph might thus be beneficial. In the future, we will support the fusion of context-compatible documents to form a valid answer to narrative query graphs, i.e., fuse content of documents that share the same context.
3.Narrative Retrieval
In the following section, we describe the process of matching narrative query graphs against documents. For this, we designed two stages: 1. A pre-processing step converts texts of documents into a graph representation. Then, these document graphs are cleaned and loaded into a structured repository. 2. A query processing step takes a user query and performs a matching within the structured repository. Finally, documents that match the user’s query are returned and visualized in a suitable frontend (see Figure 2 for a systematic overview).
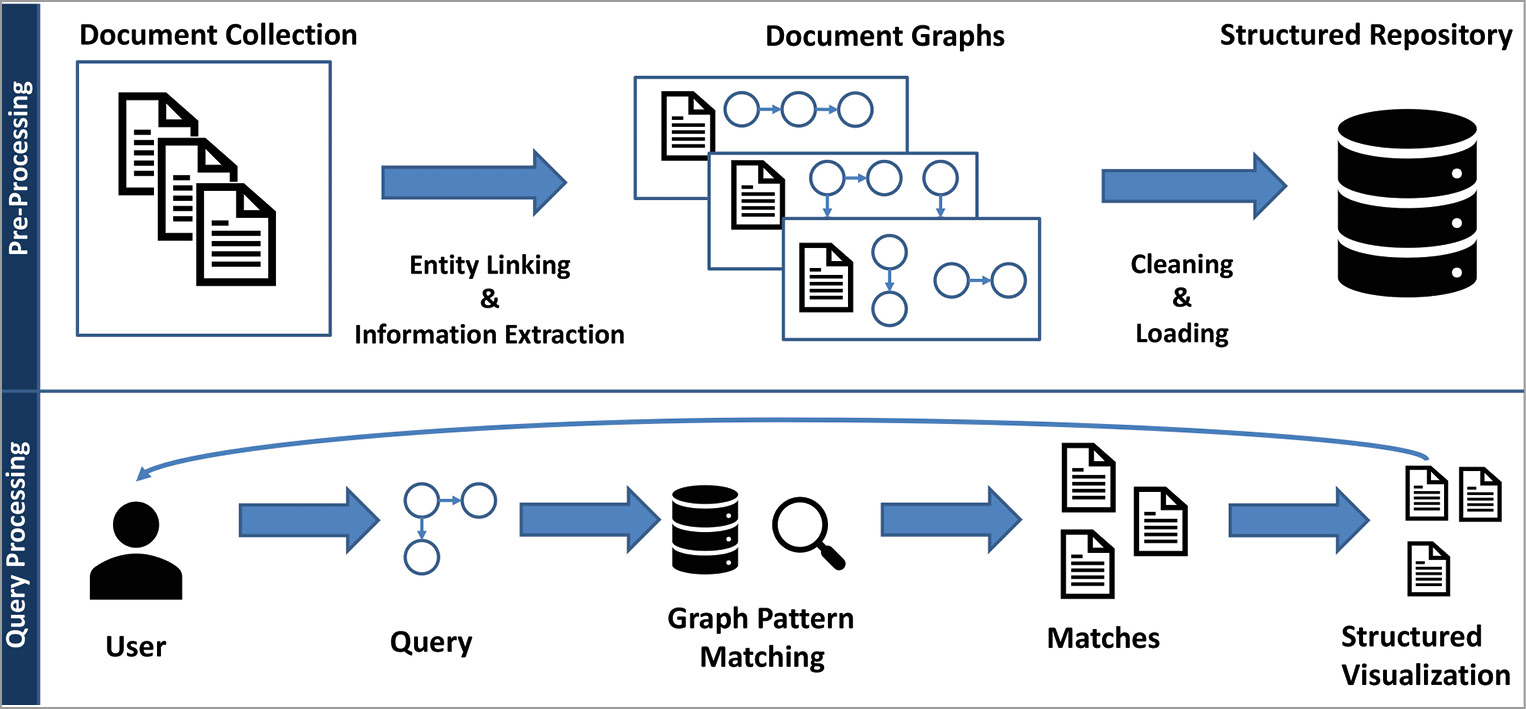
3.1Pre-Processing Texts to Document Graphs
The first step to support our narrative service is to convert natural language texts into a structured representation. Such a structured representation is required for narrative query graphs to perform an efficient and effective matching. However, information extraction from texts can usually be done in two ways: supervised and unsupervised. Supervised methods typically rely on pre-known domains and require training data to train suitable extraction models. We decided to build upon unsupervised information extraction that completely bypasses the need for training data in the information extraction phase. Therefore, we analyzed the latest open information extraction (OpenIE) tools for usage.13 OpenIE extracts structured statements based on the grammatical structure of a sentence, and it does not require domain-specific training examples. These tools tend to be precision-oriented in practice, i.e., the extractions will have a good quality, but many text statements will not be extracted. Therefore, we designed our own recall-oriented extraction method PathIE that works without supervision. PathIE requires the detection of domain-specific concepts, called entities, in the text. For this, we perform entity linking which detects pre-known entities like drugs, diseases, genes, and more in the text. For this, a domain-specific entity vocabulary must be provided. PathIE then utilizes the detected entities in the extraction phase, whereas OpenIE does not need such information. Usually, OpenIE tends to extract many unspecific concepts. In contrast, our service relies on domain-specific entities. Thus, PathIE utilizes entity linking information to extract all possible relationships between entities within a sentence. Finally, PathIE, as well as OpenIE methods, lead to extractions that may have synonymous predicates. For example, the extracted predicates treats and prevents could both describe a treatment relation. We designed an iterative cleaning procedure utilizing word embeddings and expert feedback to canonicalize these predicates. Here, a vocabulary of relations (treats, induces, etc.) and a list of synonyms must be provided to resolve the synonymous predicates. In addition to that vocabulary, our toolbox utilizes word embeddings to find more synonymous predicates automatically. Word embeddings14 represent words as vectors in a high-dimensional vector space. Here, words with a similar context, i.e., which appear in similar sentences, are closely located in the vector space. In this way, words that are synonymous or have a similar meaning can be detected. For instance, if the predicate prevents was extracted but is not contained in the relation vocabulary, the word embedding is used to find the closest match in the vector space. Here, prevents would be correctly linked to the relation treats. Finally, the pre-processing step yields cleaned document graphs.
We analyzed the advantages and disadvantages of our methods in the biomedical domain. In addition, we published all methods, including entity linking, information extraction, and cleaning, as a complete toolbox.15 The toolbox contains an entity linker, PathIE, and interfaces to the latest OpenIE techniques. All methods are designed to work nearly unsupervised and require the construction of vocabularies only. To bypass the need for entity vocabularies, we integrated support for Named Entity Recognition via the Stanford Stanza, a package of tools for natural language analysis in Python.16 In contrast to entity linking, Named Entity Recognition does not require an entity vocabulary. Usually, named entities are automatically detected by a specific set of rules, i.e., two names with a typical pattern might represent a person in the text. Modern recognition methods like Stanza are based on deep learning architectures to learn such rules from training data automatically. Stanza supports the annotation of persons, political organizations, groups, countries, works, and many more out-of-the-box. That is why Stanza is a tool that might be useful across different domains. In summary, the toolbox could be transferred to other domains, and we are looking forward to what people will be doing with it.
3.2Query Processing
Suppose that users express their information need as a narrative query graph. In that case, we must match the query against the document graphs. The pre-processing step yields cleaned document graphs. We decided to build upon a relational database for the query processing step. One might argue that NoSQL systems like graph databases or document stores might offer better performance. But on the one hand, the relational database allows to store additional metadata, e.g., titles, abstracts, publication information, and authors. On the other hand, relational databases support high-performance query processing that is sufficient for our purposes. The narrative service uses metadata to explain matches to the user, i.e., the service visualizes in which text parts the query was matched. We refer to that information as provenance information. Although there are many alternatives, we found that the relational database operates efficiently and allows us to find matches within a second in most cases.
Our system takes a narrative query graph and translates it into SQL statements for the actual query processing. Narrative query graphs might ask for several fact patterns, e.g., Metformin treats diabetes mellitus AND diabetes mellitus associated humans. Each fact pattern could be answered by querying the fact extraction table with suitable conditions. After that, the extractions must be joined by their corresponding document. This joining ensures that the whole query is matched within the document’s scope. The matching procedure uses an inverted index that maps an extracted statement to a set of supporting documents. In addition to this matching, we integrated support for ontological information. For example, diabetes mellitus is a disease with multiple forms like diabetes mellitus type 1 and diabetes mellitus type 2. If users query diabetes mellitus, they would expect to find documents that include any form of the disease. Therefore, the queries are expanded via an ontology, i.e., if users query for an entity that might have multiple subclasses, the query is expanded to find all matches. Finally, our system can compute document matches. Moreover, we also query additional provenance information to explain the returned document matches to the user, e.g., we show the sentence in which a fact was matched.
4.Narrative Prototype
In this section, we describe relevant implementation details for our service.17 The current version includes the biomedical Medline collection (PubMed).18 In the first step, all 32 million documents were processed by performing entity linking, i.e., detecting relevant biomedical entities such as drugs, diseases, genes, and more. Subsequently, the relevant pharmaceutical part of the collection was kept, i.e., documents that contain at least a single detected drug or plant family in title or abstract. This step yielded around 6.2 million documents which were transformed into document graphs and loaded into the backend.
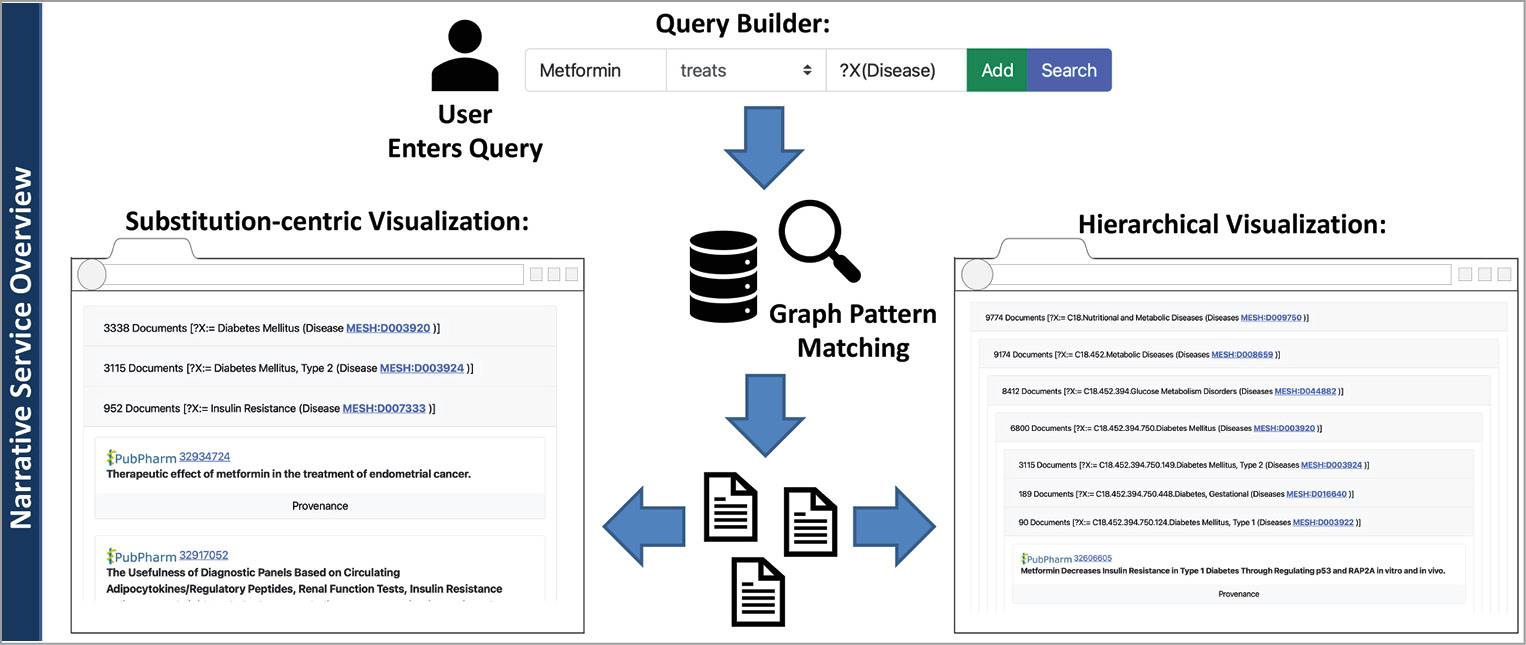
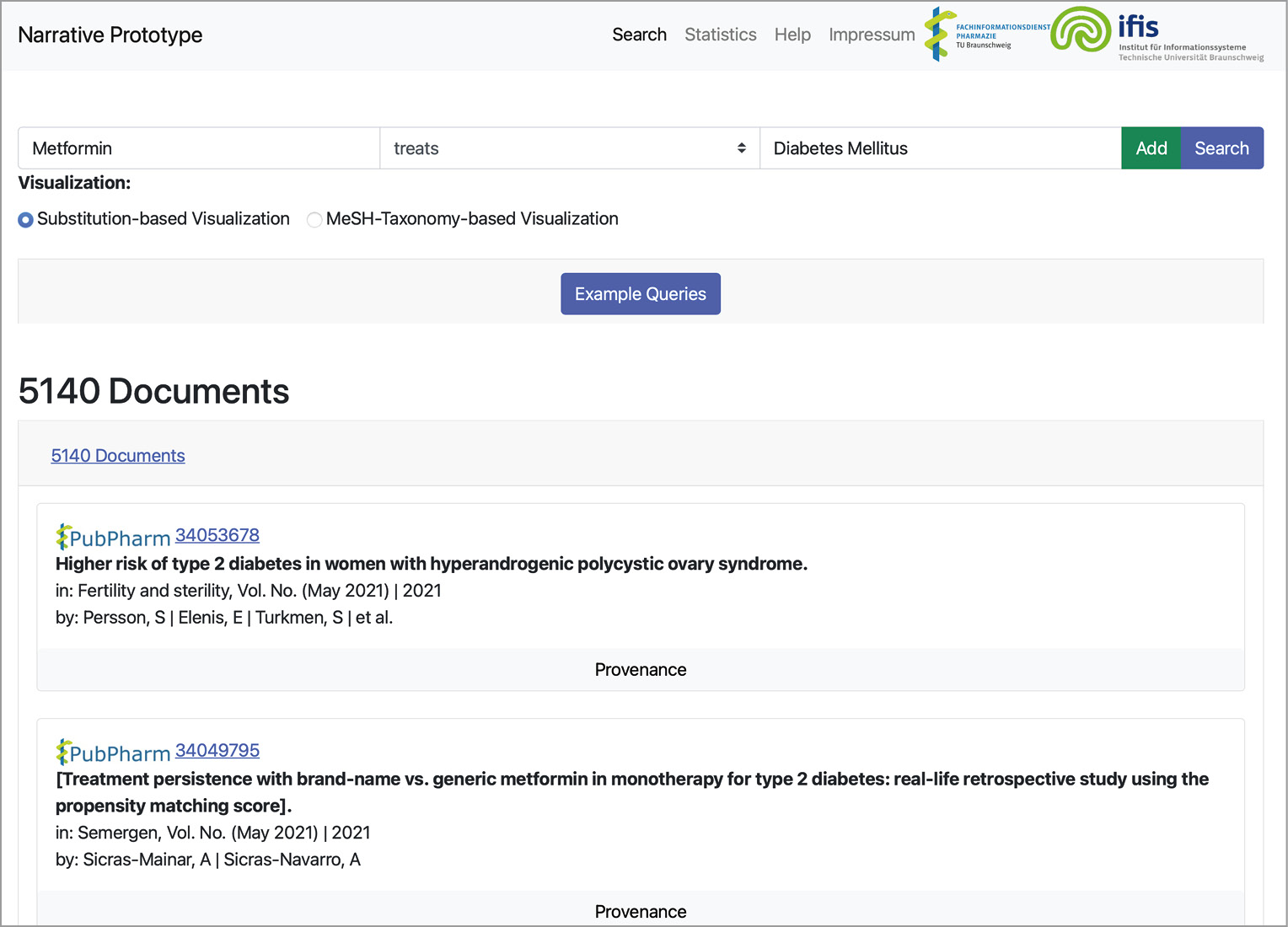
The frontend of our narrative service was implemented as a web app with the latest web techniques like Bootstrap, jQuery, and JavaScript. The backend was implemented as a REST server utilizing the Python Django server framework. And the corresponding communication between both layers was realized via REST. The prototype’s frontend consists of two essential components: the query builder and the result visualization (see Figure 3).
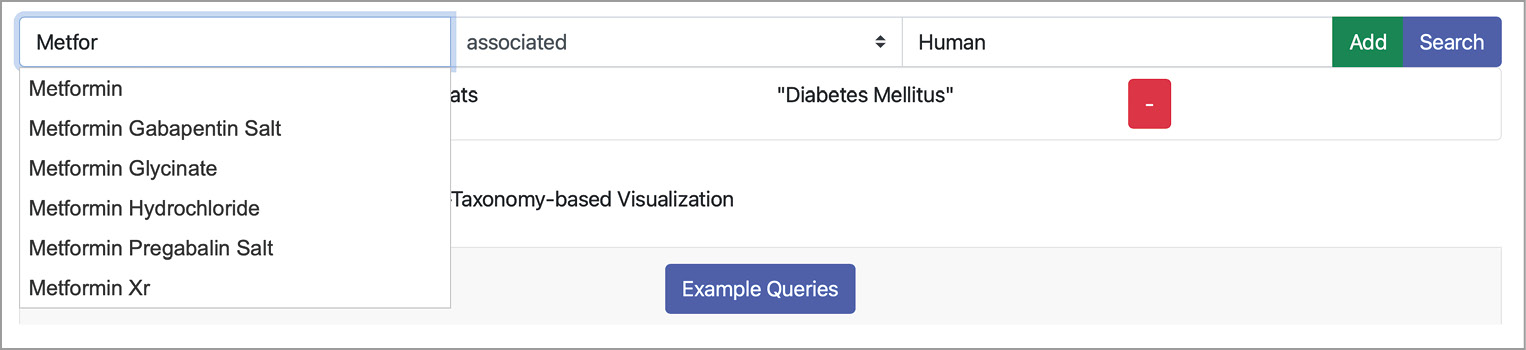
The query builder allows users to formulate a single fact pattern, i.e., subject, predicate, and object. A screenshot of the query builder is depicted in Figure 4. If a user enters a subject or an object, e.g., the drug substance Metformin, the frontend sends the typed string to the backend. The backend replies with corresponding autocompletions. Then, the frontend displays a list of possible entities as autocompletions to the user. If users complete the fact pattern, they can directly search for documents or add the fact pattern to the current query. Then, a second fact pattern can be entered, and so on. If the user finally decides to query for matches, the query is encoded as a string and send to the backend. The query is then translated into an internal format, optimized, and expanded before execution. Subsequently, the backend encodes the results in the JSON format and sends them back to the frontend.
Finally, the frontend visualizes the results (see Figure 3 and Figure 5). Here, we must distinguish between queries including variables and queries asking for specific entities. If no variable is included, the prototype visualizes the results as a list of documents. The title, journal, authors, publication year, and a provenance button are shown for each document. If users click the provenance button, a list of sentences is displayed. The service visualizes why a sentence matches a fact pattern by highlighting the sentence’s entity annotations and matched predicates (see Figure 6). Additionally, the document ID is linked to the corresponding article in the PubPharm search platform. Finally, the narrative service ranks the documents by their publication date (latest first).

Suppose that a query contains a variable, e.g., Metformin treats ?X(Disease). In that case, the service provides two different options to display the result (see Figure 3). The first option is called substitution-centric visualization. Here, documents that share the same substitution for the variable (a specific entity) are aggregated in a group. These groups are then sorted, descending by the number of contained documents. Each group is visualized as a collapsed list view. If users click on that item, the list of documents is displayed. They can also collapse the list again and browse to another group. Besides, we support a taxonomy-based visualization (hierarchical visualization). Entity types like diseases or drugs might be arranged in a taxonomy. For example, the entity diabetes mellitus type 1 is a subclass of diabetes mellitus. If the user queries disease treatments (e.g., Metformin treats ?X(Disease)), the service considers the taxonomy to visualize arranged document groups. Here, an entry might be a collapsed list of documents sharing the substitution diabetes mellitus (see Figure 3). If the user clicks on that list, more subclasses of diabetes mellitus are displayed as collapsable lists. We support this visualization to assist users in getting a better overview of the literature. Here, users may see which specific drug treats classes of diseases, and if they are interested in more information, they can expand the relevant parts of the taxonomy.
In summary, the service provides users with a structured query builder and an interactive result visualization. One of our main goals was to develop an understandable and intuitive service because we need to move from keyword-based retrieval to a new kind of query building. We already reported details about the evaluation and user studies of our service19. We performed interviews and user hands-on tests with eight pharmaceutical researchers to understand how the service supports our community. We interviewed each person at least twice and collected feedback in a qualitative evaluation. A structured query builder was the first choice in interviews with users from the pharmaceutical research community. In addition, users appreciate our interactive elements like showing provenance information or having collapsable list items. After a learning phase, users note that our service intuitively supports their daily research. In addition, users appreciate using variables in their queries to generate suitable overviews of the literature quickly. These structured overviews might be especially helpful for young researchers who are still unfamiliar with their topic.
5.Discussion
The narrative service is designed to assist in two use cases. On the one hand, we support high-precision document searches, i.e., users can precisely formulate their intended narrative of interest. On the other hand, queries with variables support entity-centric literature overviews. Due to the pre-processing step, our service lacks recall in comparison to keyword-based retrieval, for several reasons: First, entity linking and information extraction methods do not work perfectly. For example, resolving entity homonyms and finding all entity synonyms is challenging. Next, the current version of our information extraction step is restricted to single sentences. If a relationship is expressed over two different sentences, then our service cannot find it. Nevertheless, the narrative service is the first step toward a novel access path to information. For example, suppose that a scientist has some hypothesis in their mind. In that case, they could formulate their hypothesis as a narrative query graph and get precise answers to verify their hypothesis. Besides verifying a hypothesis, they could also perform a broad search, e.g., they quickly want to get an overview of the latest drug developments. Such information, especially if the information could only be found in the latest literature, would rarely be available in a curated specialized database (e.g. ChEMBL).20 The narrative service supports these use cases by considering the latest literature.
However, the narrative service was designed to assist pharmaceutical research. In addition to improving the service, we aim to share the narrative idea with different domains and communities. Therefore, we already published the pre-processing step as open source software. In the future, we will continue our community work and will share more information, code and software if possible. For example, suppose a similar service should be built for another domain. In that case, designing two different vocabularies is required. First, a vocabulary of all relevant domain-specific entities must be provided. This vocabulary will be used to detect entities mentioned in the text. The entity vocabulary is essential to resolve synonyms and to filter the text for relevant entities. Next, a vocabulary of relations must be built. In contrast to supervised extraction methods, our toolbox does not require training data for each relation. Unsupervised methods may extract various and maybe synonymous predicates, e.g., ‘prevents’, ‘treats’ and ‘cures’. The relation vocabulary will then be used to link synonymous predicates to the same relation, e.g., map the predicates ‘prevents’, ‘treats’, and ‘cures’ to the relation ‘treats’. Besides, we support the usage of word embeddings in the cleaning phase to generate more synonymous predicates for a relation automatically. We believe that our nearly unsupervised approach is an important decision to transfer this research to different domains, although unsupervised methods may lack quality. Hence, the narrative idea and the narrative service could easily be transferred to another domain. However, suitable vocabularies must be designed for this.
6.Conclusion
In conclusion, the narrative service supports two essential use cases. On the one hand, users can quickly verify a hypothesis. On the other hand, users can generate entity-centric and structured overviews of the literature. We believe that the narrative idea, i.e., making more sense of the current vast amount of data, is getting more important in the future. Especially if the collections grow as rapidly as they do nowadays, efficient and effective access paths are strongly appreciated and required by the users. The narrative model supports a more precise and structured retrieval of scientific discourses. However, our narrative retrieval is currently restricted to document collections. In the future, it might be interesting to integrate more and different kinds of knowledge repositories. As a first use case, we could improve the extraction quality by verifying extracted statements through a specialized database like ChEMBL. However, bridging the gap between different knowledge repositories such as document collections, research data, and structured databases remains a challenging task.
References
- Banko, Michele; Cafarella, Michael J.; Soderland, Stephen; Broadhead, Matthew; Etzioni, Oren: Open Information Extraction from the Web. In: IJCAI, 2007, pp. 2670–2676. Online: <http://ijcai.org/Proceedings/07/Papers/429.pdf>, last accessed 27.08.2021.
- Draheim, Christina; Keßler, Kristof; Wawrzinek, Janus; Wulle, Stefan: Die Rechercheplattform PubPharm. In: GMS Medizin - Bibliothek – Information 19 (3), 2019. Online: <https://dx.doi.org/10.3205/mbi000448>.
- Keßler, Kristof; Kroll, Hermann; Wawrzinek, Janus; Draheim, Christina; Wulle, Stefan; Stump, Katrin; Balke, Wolf-Tilo: PubPharm. Gemeinsam von der informationswissenschaftlichen Grundlagenforschung zum nachhaltigen Service. In: ABI Technik 39 (4), 2019, pp. 282–294. Online: <https://doi.org/10.1515/abitech-2019-4005>.
- Kroll, Hermann; Kalo, Jan-Christoph; Nagel, Denis; Mennicke, Stephan; Balke, Wolf-Tilo: Context-Compatible Information Fusion for Scientific Knowledge Graphs. In: International Conference on Theory and Practice of Digital Libraries. Springer, Cham, 2020, pp. 33–47. Online: <https://link.springer.com/chapter/10.1007/978-3-030-54956-5_3>, last accessed 25.08.2021.
- Kroll, Hermann; Nagel, Denis; Balke, Wolf-Tilo: Modeling Narrative Structures in Logical Overlays on Top of Knowledge Repositories. In: International Conference on Conceptual Modeling. Springer, Cham, 2020, pp. 250–260. Online: <https://link.springer.com/chapter/10.1007/978-3-030-62522-1_18>, last accessed 25.08.2021.
- Kroll, Hermann; Pirklbauer, Jan; Balke, Wolf-Tilo: A Toolbox for the Nearly-Unsupervised Construction of Digital Library Knowledge Graphs. In: ACM/IEEE Joint Conference on Digital Libraries (JCDL), 2021.
- Kroll, Hermann; Pirklbauer, Jan, Kalo; Jan-Christoph; Kunz, Morris; Ruthmann, Johannes; Balke, Wolf-Tilo: Narrative Query Graphs for Entity-Interaction-Aware Document Retrieval. In: The 23rd International Conference on Asia-Pacific Digital Libraries (ICADL), 2021.
- Mikolov, Tomas; Chen, Kai; Corrado, Greg; Dean, Jeffrey: Efficient estimation of word representations in vector space. In: ICLR Workshop, 2013. Online: <https://arxiv.org/abs/1301.3781>, last accessed 27.08.2021.
- Qi, Peng; Zhang, Yuhao; Zhang, Yuhui; Bolton, Jason; Manning, Christopher D.: Stanza. A Python Natural Language Processing Toolkit for Many Human Languages. In: Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics (ACL): System Demonstrations. ACL, 2020, pp. 101–108. Online: <http://dx.doi.org/10.18653/v1/2020.acl-demos.14>.
- Vrandečić, Denny; Krötzsch, Markus: Wikidata. A free collaborative knowledgebase. In: Communications of the ACM 57 (10), 2014, pp. 78–85. Online: <http://dx.doi.org/10.1145/2629489>.
1 Draheim, Christina; Keßler, Kristof; Wawrzinek, Janus; Wulle, Stefan: Die Rechercheplattform PubPharm. In: GMS Medizin - Bibliothek – Information 19 (3), 2019. Online: <https://dx.doi.org/10.3205/mbi000448>.
2 KEGG: Kyoto Encyclopedia of Genes and Genomes, <https://www.genome.jp/kegg/>, last accessed 25.08.2021.
3 Uniprot, <https://www.uniprot.org/>, last accessed 25.08.2021.
4 ChEMBL, <https://www.ebi.ac.uk/chembl/>, last accessed 25.08.2021.
5 DrugBank, <https://go.drugbank.com/>, last accessed 25.08.2021.
6 Keßler, Kristof; Kroll, Hermann; Wawrzinek, Janus; Draheim, Christina; Wulle, Stefan; Stump, Katrin; Balke, Wolf-Tilo: PubPharm. Gemeinsam von der informationswissenschaftlichen Grundlagenforschung zum nachhaltigen Service. In: ABI Technik 39 (4), 2019, pp. 282–294. Online: <https://doi.org/10.1515/abitech-2019-4005>.
7 Kroll, Hermann; Pirklbauer, Jan; Balke, Wolf-Tilo: A Toolbox for the Nearly-Unsupervised Construction of Digital Library Knowledge Graphs. In: ACM/IEEE Joint Conference on Digital Libraries (JCDL), 2021.
8 Toolbox for the nearly unsupervised extraction of knowledge from texts, <https://github.com/HermannKroll/KGExtractionToolbox>, last accessed 27.08.2021.
9 Kroll, Hermann; Pirklbauer, Jan; Kalo, Jan-Christoph; Kunz, Morris; Ruthmann, Johannes; Balke, Wolf-Tilo: Narrative Query Graphs for Entity-Interaction-Aware Document Retrieval. In: The 23rd International Conference on Asia-PacificDigital Libraries (ICADL), 2021.
10 Vrandečić, Denny; Krötzsch, Markus: Wikidata: a free collaborative knowledgebase. In: Communications of the ACM 57 (10), 2014, pp. 78–85. Online: <http://dx.doi.org/10.1145/2629489>.
11 Kroll, Hermann; Nagel, Denis; Balke, Wolf-Tilo: Modeling Narrative Structures in Logical Overlays on Top of Knowledge Repositories. In: International Conference on Conceptual Modeling. Springer, Cham, 2020, pp. 250–260. Online: <https://link.springer.com/chapter/10.1007/978-3-030-62522-1_18>, last accessed 25.08.2021.
12 Kroll, Hermann; Kalo, Jan-Christoph; Nagel, Denis; Mennicke, Stephan; Balke, Wolf-Tilo: Context-Compatible Information Fusion for Scientific Knowledge Graphs. In: International Conference on Theory and Practice of Digital Libraries. Springer, Cham, 2020, pp. 33–47. Online: <https://link.springer.com/chapter/10.1007/978-3-030-54956-5_3>, last accessed 25.08.2021.
13 Banko, Michele; Cafarella, Michael J.; Soderland, Stephen; Broadhead, Matthew; Etzioni, Oren: Open Information Extraction from the Web. In: IJCAI, 2007, pp. 2670–2676. Online: <http://ijcai.org/Proceedings/07/Papers/429.pdf>, last accessed 27.08.2021.
14 Mikolov, Tomas; Chen, Kai; Corrado, Greg; Dean, Jeffrey: Efficient estimation of word representations in vector space. In: ICLR Workshop, 2013. Online: <https://arxiv.org/abs/1301.3781>, last accessed 27.08.2021.
15 Kroll, Hermann; Pirklbauer, Jan; Balke, Wolf-Tilo: A Toolbox for the Nearly-Unsupervised Construction of Digital Library Knowledge Graphs. In: ACM/IEEE Joint Conference on Digital Libraries (JCDL), 2021.
16 Qi, Peng; Zhang, Yuhao; Zhang, Yuhui; Bolton, Jason; Manning, Christopher D.: Stanza: A Python Natural Language Processing Toolkit for Many Human Languages. In: Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics (ACL): System Demonstrations. ACL, 2020, pp. 101–108. Online: <http://dx.doi.org/10.18653/v1/2020.acl-demos.14>.
17 Narrative Prototype, <http://www.pubpharm.de/services/prototypes/narratives/>, last accessed 25.08.2021.
18 PubMed, <https://pubmed.ncbi.nlm.nih.gov/>, last accessed 25.08.2021.
19 Kroll, Hermann; Pirklbauer, Jan; Kalo, Jan-Christoph; Kunz, Morris; Ruthmann, Johannes; Balke, Wolf-Tilo: Narrative Query Graphs for Entity-Interaction-Aware Document Retrieval. In: The 23rd International Conference on Asia-Pacific Digital Libraries (ICADL), 2021.
20 ChEMBL, <https://www.ebi.ac.uk/chembl/>, last accessed 25.08.2021.